This blog post is provided by Emil Ellegaard Thomassen and Philip Francis Thomsen and tells the #StoryBehindThePaper for the paper “Environmental DNA metabarcoding reveals temporal dynamics but functional stability of arthropod communities in cattle dung” which was recently published in the Journal of Animal Ecology. In their study, they use eDNA analysis to give a detailed picture of species communities in the dung pads at different timepoints.
From the perspective of a small insect, a fresh dung pile dropped by a cow, or another large mammal represents a rich pool of resources, but one which is only available relatively briefly. Among dung-associated insects competing for this resource, several mechanisms have evolved for rapidly finding the dung, as well as subsequently surviving and reproducing in this substrate. Fascinating examples include the antennas of scarab beetles (Fig. 1),refined for detecting scents emitted from fresh dung and even for discriminating between dung types, and the leg morphology of dung-digging beetles optimised for digging through dung and the soil below.

Once the dung is dropped, the race for the resource begins. The flies are the fastest, some arriving almost before the dung hits the ground, followed by the dung beetles of which some dwell in the dung pats, while others dig tunnels below, or transport dung balls across vast distances to their burrows.
Investigating community succession with environmental DNA
The exact timing of arrival and turnover of species through time after the dung has been deposited, is however not fully known, especially when it comes to small, understudied groups of insects and other arthropods. Another important question is to what extent the functions performed by dung-associated species are stable, and resilient to species losses? Specifically, are there redundancy of functionality such that many co-occurring species in the dung perform similar functions? Luckily, recent developments in DNA technologies have introduced new means of investigating diverse biological communities in a non-invasive, and resource-efficient way by extracting DNA from environmental samples(eDNA).
We investigated the development of arthropod communities through time, in a natural experiment, where we collected large quantities of fresh cattle dung, mixed it, and divided it into smaller pools of equal size which we placed in the study area. These pools represented freshly deposited dung pads, ready to be colonised by dung-associated insects. We then collected samples for eDNA analysis at several timepoints after deposition and obtained a detailed picture of the species communities in the dung pads at different timepoints (Fig. 2).
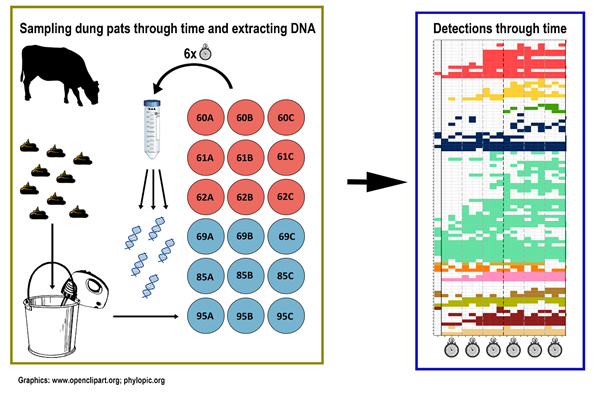
Timing of arrival and methodological caveats
We identified species that were mainly present at the initial sampling points after the dung has been deposited, such as the flies Hylemya vagans, Scathophaga stercoraria, and Neomyia cornicina, some that were present through most of the experimental period (e.g., the dung beetle Aphodius heamorrhoidalis and the fly Musca autumnalis), and some that were only present at the last timepoints (e.g., the beetles Cryptopleurum minutum, Philonthus albipes & Ptilium exaratum). The late arrivals especially included species from the functional groups representing higher trophic levels, such as predatory beetles, and fungivorous beetles and flies.
Our results show that environmental DNA can be a valuable tool for detecting changes in communities of dung-associated arthropods through time, and much information on small elusive species that are hard to sample and identify morphologically can be obtained by this approach.
The method does have some limitations. Seeing our results, we were very puzzled about why the only detected dung-digging beetle was Onthophagus similis, though several other species from this functional group were present in the study area. This includes the dor beetles (Anoplotrupes stercorosus and Trypocopris vernalis) and other species from the genus Onthophagus such as O. fracticornis (Fig. 3).

The dung dwellers (genus: Aphodius) were more commonly detected in our samples, and we think this is a result of the differences in the biology and life cycles of these species, where larvae of the dung-dwellers are present in the dung. This lifestyle means they have a higher level of activity within the dung through time, potentially leaving more DNA traces behind. The dung-digging species, who are digging through the dung and transporting dung to the burrows where the larvae develop might leave significantly less DNA traces, and consequently decrease the probability of detection by eDNA-based identification.
Another property of eDNA-based identification is that DNA is preserved in the dung for some time after the organism itself has disappeared. This is a disadvantage when looking at fine-scale changes over time, as it results in detection of species at timepoints after they have disappeared. Our data reveals this, as we find much more examples of species associated with late compared to early timepoints, but when we looked at the relative proportions of DNA sequences in the samples, we were able to detect a signal of early activity for some species whereafter the DNA started to degrade. Thus, looking at patterns of DNA degradation in the samples could help identify peaks in activity, which are not evident from presence-absence data only.
Prospects for investigations of dung-associated communities
So, what have we learned? Importantly, our study found that the groups especially important for degradation of dung were highly diverse, and constantly represented through time, with some species present from the beginning, and others arriving at later timepoints. This suggests that functional stability exists, and the system might be functionally resilient to loss of dung-degrading species. Through our study, we show that eDNA-based methods can be highly useful for investigating dung-associated arthropod communities over time, but importantly, there are species groups which are missed by this approach. This calls for integrative approaches, and we suggest that thorough understanding of arthropod succession in cattle dung could be obtained by combining eDNA-based identifications with traditional morphology-based observations, and other next-generation monitoring approaches such as computer vision for image identification.
Read the paper
Read the full paper here: Thomassen, E. E., Sigsgaard, E. E., Jensen, M. R., Olsen, K., Hansen, M. D. D., & Thomsen, P. F. (2024). Environmental DNA metabarcoding reveals temporal dynamics but functional stability of arthropod communities in cattle dung. Journal of Animal Ecology, 00, 1–19. https://doi.org/10.1111/1365-2656.14119
